Week 1
7/23/12
After a day of orientation, we divided up into lab groups based roughly on our experience with different techniques/fields. I'm with Annie and Courtney - we're the only lab group that has to look across the whole lab to look out the window and see the ocean.7/24/12
Following a lecture on general invertebrate anatomy (of gastropods, molluscs, crustaceans), we spent the morning's lab working on dissections.7/25/12
| Armina digestive gland being placed into a histology cassette |
7/26/12
DNA Extractions from Armina TissueDNA extraction from the tissue samples taken yesterday from Armina, accession number 12-1-6 for the sample I extracted. Followed protocol exactly but at the point of pipetting the sample into the spin column, made the following mistake. Instead of pipetting the full sample into the column, took 200 uL into one spin column, so did another column with the rest of the sample. Used the same amount of buffer for intermediate steps with the column and then combined DNA extractions after elution. DNA extraction stored in the freezer/fridge (?) until the afternoon, at which time the following PCR was run.
PCR Master Mix Calculations (originally done in excel)
| Two Primer Sets (generic) Master Mix |
|
8 |
||
| uL/Reaction |
uL/8 Reactions |
|||
| Immomix 2x |
12.5 |
100 |
||
| BSA (10 mg/ml) |
1.5 |
12 |
||
| Fwd primer (10 um) |
0.8 |
6.4 |
||
| Rev primer (10 um) |
0.8 |
6.4 |
||
| sH2O |
7.4 |
59.2 |
||
| Template |
2 |
16 |
23 |
|
| Total |
25 |
200 |
||
| WS-RLO cPCR Master Mix |
|
8 |
||
| Reagent |
[Stock] |
[End] |
Per reaction (uL) |
Per 8 reactions (uL) |
| 5 X Buffer |
5 X |
1 X |
4 |
32 |
| MgCl2 |
25 mM |
1.5 mM |
1.2 |
9.6 |
| BSA |
10 mg/ml |
400 ng/ml |
0.8 |
6.4 |
| H2O |
11.08 |
88.64 |
||
| dNTP's |
10 mM |
200 uM |
0.4 |
3.2 |
| RA 3-6 |
100 pmol/ml |
0.5 uM |
0.1 |
0.8 |
| RA 5-1 |
100 pmol/ml |
0.5 uM |
0.1 |
0.8 |
| Taq |
5 U/ul |
1.6 U |
0.32 |
2.56 |
| Template |
2 |
16 |
||
| Total |
20 |
160 |
||
| 1 |
2 |
3 |
4 |
5 |
6 |
| Negative Control |
Blank Control |
CC 12-1-6 |
GS 12-1-6 |
2010 Sample (Annie) |
Positive Control |
Additional Armina Dissection with Maya
- 12-1-7, ~54 mm, ~3.4 g
- Counted approximately 18 lesions on the dorsal surface, ranging from point lesions to multi-focal lesions
- Transverse sections for histology; one section of digestive gland tissue and another of the lesions themselves
- Two tissue samples for DNA extraction, stored in ethanol at room temperature
- Placed the histology cassette in Davidson's invertebrate fixative at approximately 5:30 pm
7/27/12
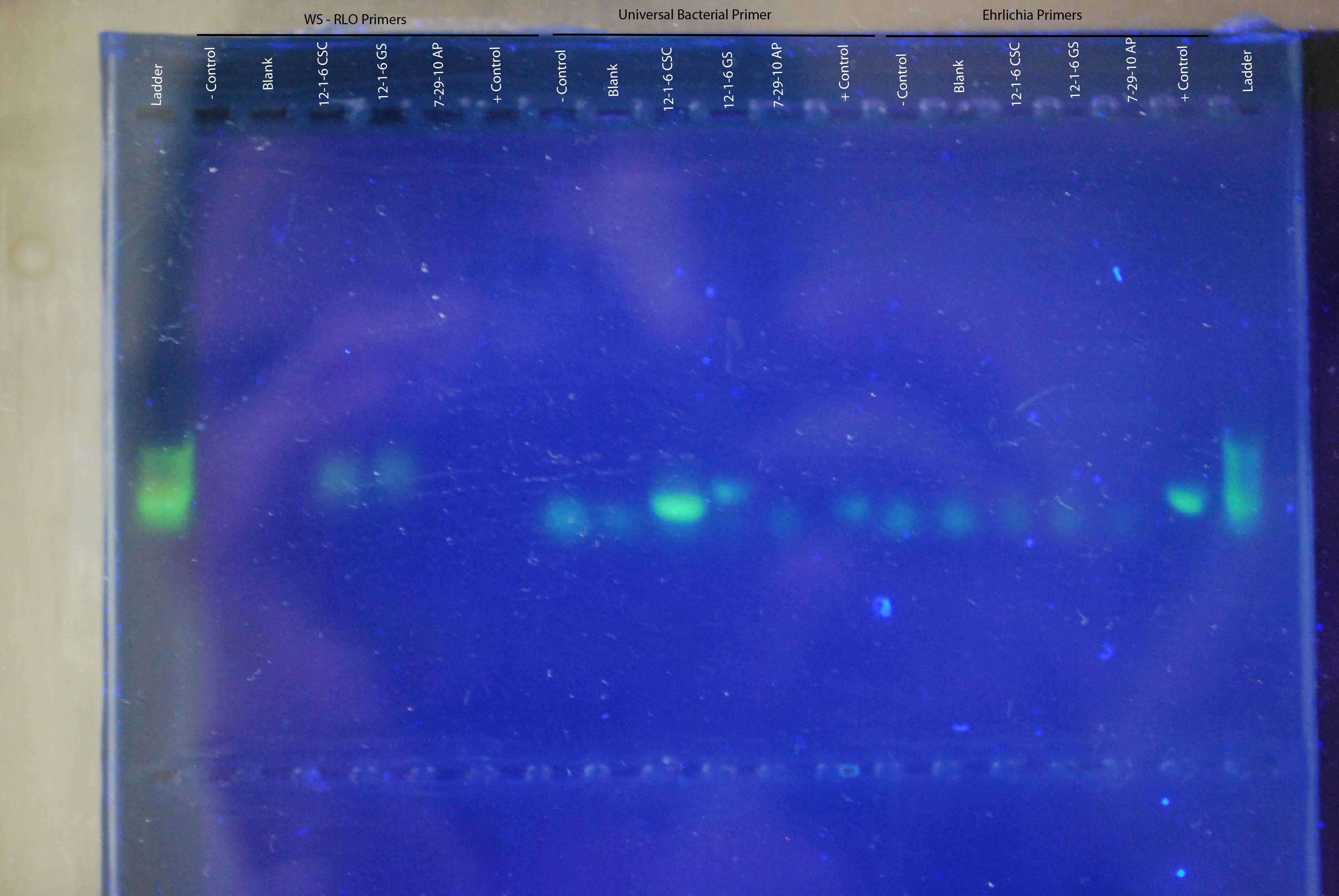
Gel of Yesterday's PCRInstead of using ethidium bromide (a very bad carcinogen) we will be using Cybersafe (a not so bad carcinogen) to stain the gel, intercalate with the DNA and visualize the amplified product.
Visualization of gel using transilluminator. Gel product shown in the image below, with primer sets and samples labeled. No positive control showed up for the WS-RLO primers, but both the universal bacterial and ehrlichia primers showed a band at the positive control. Universal bacterial primers showed a band for the 12-1-6 CSC, which was a piece of digestive gland tissue isolated from an Armina dissected in 2012. Other bands that are faint and don't line up with the positive control are likely primer dimers.
Week 2
8/3/12
Team Crosson Protocol for Inoculations (written by Sonia)
Day 1-2
- Grow up Laby cultures at desired temperatures
- Cut a putatively healthy blade of eelgrass into pieces of 2cm (possibly more). Leave in the seatable to allow cuts to heal.
- Vortex the Laby cultures.
- Use a hemocytometer to gain some indication of culture densities.
- Based on the density of the full culture, make serial dilutions of 10x or 100x. OR pellet the Laby cells and resuspend to desired density in seawater.
- Sterilize the eelgrass pieces and place them on a petri dish (with some seawater?). Perhaps scratch or provide another form of mechanical stress as a point of entry for the Laby.
- Squirt a set volume of resuspended/ diluted Laby culture onto each blade piece. Control set would receive a squirt of seawater with no Laby.
- Allow to grow/ infect.
- Record presence/ absence of lesions. Use imaging software to analyze the size of lesions in each treatment.
Team Harvell Protocol for Inoculations (written by Sonia, adapted from [Muhlstein et al. 1988])
Day 1
- Place plants to be inoculated into a 2-liter Erlenmeyer flask (or hollow plastic tube) and allow to acclimate for 48 hours. Literature conditions:
- Aerated artificial seawater at 36% salinity OR 0.45µM Millipore-filtered natural seawater at 36% salinity.
- 300 µE/(m*s) of illumination on a 14h light : 10 h dark cycle
- Constant temperature of 17 C
- Autoclave small (1-cm) pieces of healthy eelgrass in seawater. Place the pieces on an agar surface containing Labyrinthula. (In the paper, yeast acted as a food source for the Labyrinthula cultures. The authors report that the Labyrinthula would grow a! way from the yeast after 2-3 days.)
- Allow live plants to continue to acclimate.
- Allow Labyrinthula to invade the sterilized pieces of eelgrass.
- Once the sterilized eelgrass leaves have been invaded, perform the inoculations within 2-4 days:
- Clamp a single invaded leaf piece to a healthy, living eelgrass shoot 1-2 c! m above the sheath.
- Monitor every 3-4 hours for the first 24 hours (perhaps excessive for our purposes), then daily for the appearance of disease symptoms.
- Control plant: Clamp an autoclaved leaf from a clean petri dish (no Labyrinthula).
- Use imaging software to compare lesion sizes in inoculated and control plants.
- Optional: Re-isolate Labyrinthula from the lesions by plating on SSA.
Week 3
8/3/12
Week 4
8/13/12
PM Lab: Learning how to play with BLAST and doing things with "My favorite gene"- Download UniProtKB/SwissProt in FASTA format at http://www.uniprot.org/downloads
- Applications -> blast
- CL_10 -> Users -> Shared -> EIMD_blast -> db -> uniprot_sprot.fasta.gz -> uniprot_sprot.fasta
Terminal (how to access BLAST through the terminal)
- ls - list current directory contents
- pwd - current location
- cd /Applications/blast - changes directory to the blast folder, which has the same effect as typing cd and dragging in the desired folder
- cd /bin - changes directory into the bin folder
- cd /Applications/blast/bin -- fhlguest$ ./makeblastdb -help
- within the bin directory, do: ./makeblastdb -in /Users/Shared/EIMD_blast/db/uniprot_sprot.fasta -out /Users/Shared/EIMD_blast/db/uniprot_sprot -dbtype prot
- ./blastx -query /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta -db /Users/Shared/EIMD_blast/db/uniprot_sprot
How to run an actual BLAST from the command line for the QPX data:
- ./blastx -query /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta -db /Users/Shared/EIMD_blast/db/uniprot_sprot -max_target_seqs 1 -outfmt 6 -evalue 1E-5 -out /Users/Shared/EIMD_blast/out/qpxblastswissprot
8/14/12
AM Lab: QPX Data AnalysisWe received the first batch of data off the Illumina Hi-Seq machine.
With regard to QPX- three libraries were sequenced
- DNA
- mRNA with culture grown at 10C
- mRNA with culture grown at 21C
You can see the raw data (or download) from the DNA library @
http://aquacul4.fish.washington.edu/~steven/armina/filtered_QPX_DNA_GTGAAA_L001_R1.fastq.gz
de novo assembly using CLC workbench. There are other types of software to do this with, including Trinity (?), Geneious, Newbler. Assembly uses the "Greedy algorithm", and we can specify the stringency requirements depending on species, HTS machine. Can also use paired-end sequencing to overlay a spatial component onto sequence data, placing the short reads in relationship to each other.
Map reads back onto the assembled contigs to generate an RNA-seq file. Set parameters s.t. number of mismatches is 2, and specify the number of times a read can go to multiple contigs. Tab delimited file will return data including coverage, gene length, etc. and RPKM (which normalizes for library size:
"Feature ID" "Expression values" "Gene length" "Unique gene reads" "Total gene reads" "RPKM"Use CLC to compare the two QPX treatment libraries in an "experiment" to generate a file that can be cut down into fold-change difference between the two: CLC Fold-Change Results
Proceed by BLASTing the transcriptome (FULL transcriptome) that was assembled from the 10 and 21 degree C libraries. So, to do this we are going to do the following.
- Open /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta in TextWrangler
- Find "1 Contig " replace all "1_Contig_"
- Command line: on local computer, find /Applications/blast/bin
- ./blastx -query /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta -db /Users/Shared/EIMD_blast/db/uniprot_sprot -out /Users/Shared/EIMD_blast/out/QPXSwissProtBLAST -outfmt 6 -evalue 1E-20 -max_target_seqs 1 -num_threads 2
- #CPU use: num_threads (1 or 2)
- #what you want to BLAST: -query
- #database to blast against: -db
- #location to place output file: -out
- #output format: -outfmt [number 6 = tab delimited]
- #stringency: evalue [1E-20]
- #hits per contig: -max_target_seqs #
PM Lab:
Upload "fold-change" file to Galaxy. Download the transcriptome->SwissProt mapping file.
Join tables from transcriptome, gene expression change table,
- Open Galaxy (Penn State).
- Upload file with headers into Galaxy, no changes to format of uploading
- Text Manipulation -> Convert Delimiters to Tabs -> Pipes to Tabs
- Download databases from SR320's Galaxy history. In this case we pull "SPID and Description" into Galaxy
- Join two tables, join column 4 from the annotated BLAST file (imported from Sonia with headers) with column 1 of the SWPID file, change all settings to "Yes"
- Result is an annotated transcriptome
- Assign function: import SwissProt "associations" file from Sr320's history and join to the table constructed above
- GO Terms: cellular component, molecular function, biological process
- Join the GO Association table with the "GO to GO Slim"
With the GO Slim Association Table
Import file, TAB delimit, join with SPID and description, join with Swiss + Associations, join with
8/15/12
AM Lab:- Download QPX_Experiment_UniqueReads_simple.txt and open it in Excel
- Find counts of down and upregulated genes
- Comparison of counts: 4783 are downregulated at higher temperature (21 C), 112 are upregulated at higher temperature (21 C)
Enrichment Analysis
- Want two files: SwissProt for entire transcriptome, SwissProt for the subset.
- Contig name and Swiss Prot ID
Gene Set enrichment analysis on DAVID
- In DAVID, Start Analysis. Upload two lists, starting with a list of the DE file SPIDs, Select the Identifier as "UniProt Accession".
- For the DE gene list, it's the gene list. For the transcriptome, it's the background. Convert to DAVID format. Convert all to proper format.
- Background gene list: Upload in the same way and specify which background to use for your DE genes.
- "Annotation Summary Results" - what can you do with the analysis
- GO Terms, GOFAT Results
- For the biological process GO terms, want to download a tab delimited text file and convert it into an excel file.
- Delimit via ~ to pull GO number from GO function
- In REVIGO pull out p value less than 0.05, can also use Cytoscape to analyze files
- There's also an R Script option
8/16/12
Metagenomic Analyses- Download files from sr320's dropbox, run with code: ./blastn -num_threads 3 -out /Users/admin/Dropbox/Cluster/BLAST/output/BlackAb_NWS_nt_node2_blastout_hit10 -db /CLC_blastdatabases/nt -query /Users/admin/Dropbox/Cluster/BLAST/query/BlackAb_NWS_assembly1.fa -outfmt 6 -max_target_seqs 4 -evalue 1E-20 -task blastn
- Galaxy -> Metagenomic Analyses
- Fetch taxonomic representation, wants GI numbers, separate from pipes
- Column 3 is GI number, name is in column 1
PM Lab
8/17/12
QPX Transcriptome to Genome Blast, using genome contigs with 1000 bp lengthBLAST2GFF
- want to take a BLAST transcriptome to genome to GFF (file format) that will then allow visualizing mapping of the genome
- BLAST file can't have headers, and use the 2_Blast2Gff.pl script in the sr320 evernote notebook
- dhcp160:Blast2GFF fhlguest$ perl ./2_Blast2Gff.pl -i /Users/fhlguest/Desktop/Siegmund/Friday/qpxblasttranscriptome_headed_run -o /Users/fhlguest/Desktop/Siegmund/Friday/qpxblasttranscriptome.gff -d "QPX_Genome" -p EXON -s "something"
8/18/12
Why share data?- building on other people's work
- you don't know everything, other people can help,
- your work can be used by others for things you're not thinking about right now
- real-time collaboration
- obligation to share because your research is funded by federal taxpayers
- sharing builds teamwork, secrets build resentment
- Low-risk, potential high-reward: proposals from graduate students are great (eg. NSF GRFP), but the person with the better outreach gets the heads up
- Crowd-funding: solicit funds from the general public on scifund (eg. SciFund Challenge, Kickstarter, Jared
- tools to share science: Blogging, most people get their news from facebook/social media, Twitter,
- Lab youtube, tumblr -> facebook, project derived blogs (eg. Ocean Acidification)
- Lab webcams, student website
- Figshare: datasets, unfunded grant proposals, everything is time-stamped, attribution to what you did
8/19/12
BLAST2GFF trying to map nonsynonymous SNP mutations to the QPX genome- dhcp155:~ fhlguest$ cd/Volumes/GSiegmund\ External\ Hard\ Drive/Blast2GFF
- -bash: cd/Volumes/GSiegmund External Hard Drive/Blast2GFF: No such file or directory
- dhcp155:~ fhlguest$ cd /Volumes/GSiegmund\ External\ Hard\ Drive/Blast2GFF
- dhcp155:Blast2GFF fhlguest$ perl ./2_Blast2Gff.pl -i/Volumes/GSiegmund\ External\ Hard\ Drive/Siegmund/Sunday/Physo3AA_QPXv017_node2_blastout -o /Volumes/GSiegmund\ External\ Hard\ Drive/Siegmund/Sunday/Physo3AA_QPXv017_node2_blastout.gff -d "QPX_Genome" -p EXON -s "something"
Once the GFF file was generated, it was uploaded to Galaxy, specifying the file format to be GFF. Once uploaded, the file was converted to a BED format and uploaded to the QPX genome track.
8/21/12
- Construct new nucleotide database from contigs of 100 bp or longer, in order to more fully describe the genome
- ocean5:bin fhlguest$ ./makeblastdb -in /Users/Shared/EIMD_blast/db/QPX_v015.fasta -out /Users/Shared/EIMD_blast/db/QPX_v015 -dbtype nucl
- ./blastn -query /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta -db /Users/Shared/EIMD_blast/db/QPX_v015 -outfmt 6 -out /Users/Shared/EIMD_blast/out/QPXblasttranscriptome -evalue 1E-25 -max_target_seqs 1
- This maps the transcriptome (version 1, not sure what the other, newer versions are?) to the 100 bp+ genome
- Saved the QPXblasttranscriptome_headed under external hard drive, Siegmund, Genome
- DON'T add headers to the file
In GALAXY
- Convert the transcriptome GFF to a BED file
- getorf (see Hard Drive for parameters)
8/21/12
- Methylation specific restriction digest
- HpaII and MspI are isoschizmers, have same recognition sites and HpaII can't cut if a cytosine is methylated but MspI that can - so you get a difference
- cut by MspI will produce a smear on the band
- HpaII will get more longer bands where methylation can't happen
- Bioinformatics approach
- methylated cytosines are highly mutable
- methylated regions of DNA are depleted of CpG dinucleotides over evolutionary time (CpG to TpG)
- Use a CpG O/E = CpG observed/CpG expected
- would expect to see 4 based on GC content - will have low ratio, but with a low ratio you then expect methylation, with a high ratio you expect unmethylation
- Average ratio for each biological process to predict the methylation status of each group.
- Genome-wide methylation
- MBD-seq methyl-binding domain isolated genome sequencing
- map enrichment level in MBD library versus CpG O/E ratio
- Bisulfite sequencing
- Reduce genome with MBD, create an Illumina library, bisulfite conversion
- bisulfite converison turns any unmethylated cytosine to a thymine
- Data analysis
- BSMAP software - map back to de novo contigs and to characterized genes in Pacific Oysters
- Visualization:
- Galaxy: add tracks of exons, CG sites, MBD-mapped reads and with high enough coverage you can map all methylated cytosines
- regions that are heavily methylated
- map transcriptome back to genome
- blastn ESTs - covers a lot of the exons, mRNA is transcribed that is transcribed but not expressed in current genome.
- Nanostring: nCounter
- Screen multiple sample types, multiple loci. See that we get different type of methylation across tissue types
- Targeted bisulfite sequencing
- MBD-seq methyl-binding domain isolated genome sequencing
- More SNPs in less methylated genes!!!! (???)
- Inducibles
- Increased variation in environmental response - a stressor could be disease
- alterative splicing
- sequence variation
- transient methylation
- POTENTIAL trade-offs between methylation and genetic diversity? ability to respond via methylation sites or increased genetic diversity, more potential for C->T
- Virulence in pathogen vs. host immune system, host and pathogen mechanism for response to the other
- Increased variation in environmental response - a stressor could be disease